PhyloNext results exploration and analysis¶
- Manually configure static map using the output in GeoPackage format
- Explore the results using Biodiverse program
- Prepare citable derived dataset with unique DOI
Constructing maps using the output in GeoPackage format¶
GeoPackage is an open format to store geospatial information.
It allows to store vector features (e.g., H3 polygons) along with their attributes (e.g., diversity estimates).
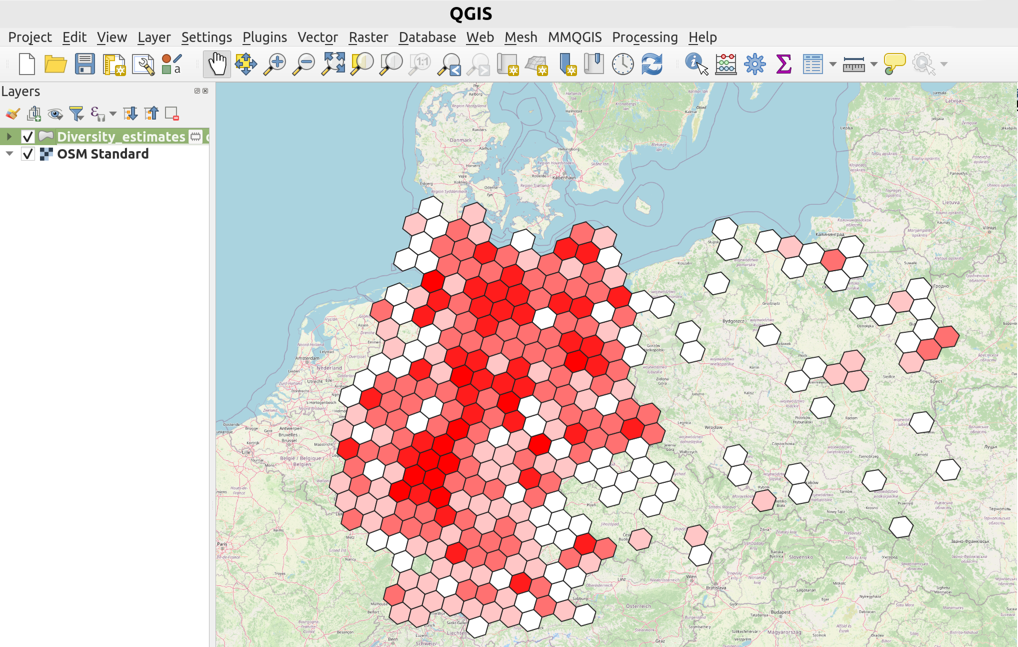
As one of the outputs, PhyloNext provides 03.Plots/Diversity_estimates.gpkg file,
which could be easily exported and visualized in your favorite GIS software.
For example, you may use a free and open source application QGIS to open a GeoPackage file.
Alternatively, it is possible to open the GeoPackage file using R or Python. Below, there are two basic examples on how to load the file. Depending on the specific requirements of the analysis, you may need to perform additional steps such as spatial transformations, querying, or more complex plotting.
In R using the sf package¶
The sf package in R is a powerful tool for handling spatial data.
It provides straightforward mechanisms for reading, writing, and manipulating spatial data in various formats including GeoPackage.
# Install sf package if it's not already installed
if (!requireNamespace("sf", quietly = TRUE)){ install.packages("sf") }
# Load the sf package
library(sf)
# Load the GeoPackage file
div <- st_read("03.Plots/Diversity_estimates.gpkg")
# Plot the data
plot(div)
In Python using the Fiona and geopandas packages¶
Fiona is used for reading and writing spatial data files,
and GeoPandas extends the datatypes used by pandas to allow spatial operations on geometric types.
# Import packages
import fiona
import geopandas as gpd
import matplotlib.pyplot as plt
# Load a GeoPackage file
div = gpd.read_file("03.Plots/Diversity_estimates.gpkg")
# Plot the data
div.plot()
plt.show()
Results exploration with Biodiverse program¶
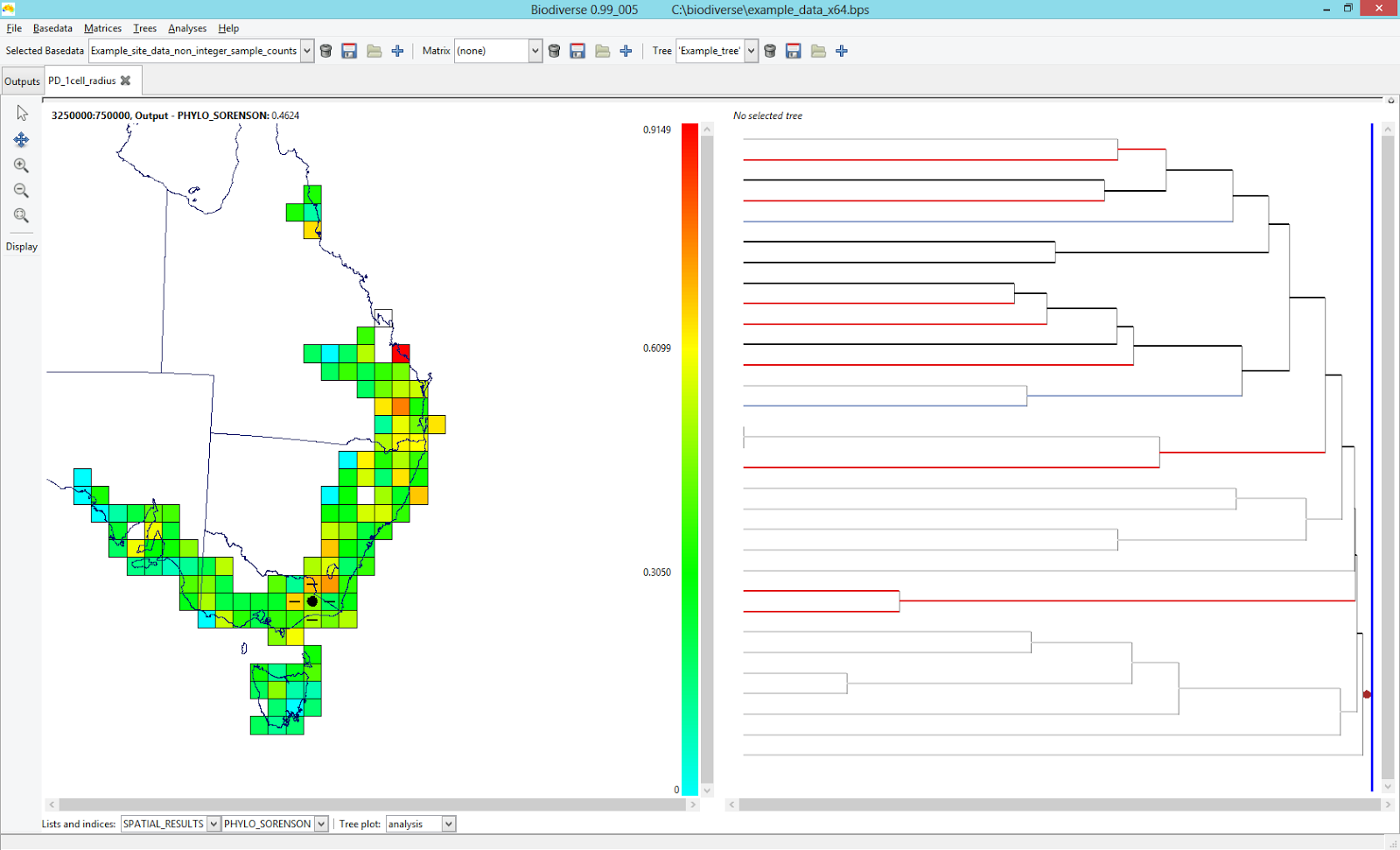
Biodiverse is a program for the spatial analysis of diversity which performs all the hard work for PhyloNext.
By default, PhyloNext output in Biodiverse format is stored in the 02.Biodiverse_results/Biodiverse.bds file.
This file could be exported to Biodiverse and visualized using graphical user interface.
Biodiverse installation
Please follow this guide to install Biodiverse.
For more details on visualization of spatial analysis results on the phylogenetic tree, see these two blog posts ( 1 and 2 ) by Shawn Laffan.
Derived datasets¶
Derived datasets are citable records (with a unique DOI)
representing GBIF-mediated species occurrences that has been filtered significantly.
To create a derived dataset, PhyloNext provides a table with a list of the GBIF datasets (defined as datasetKey) from which the data originated.
User can register new derived dataset at GBIF.org following this link:
https://www.gbif.org/derived-dataset
For additional information please see this GBIF blog post by Daniel Noesgaard.